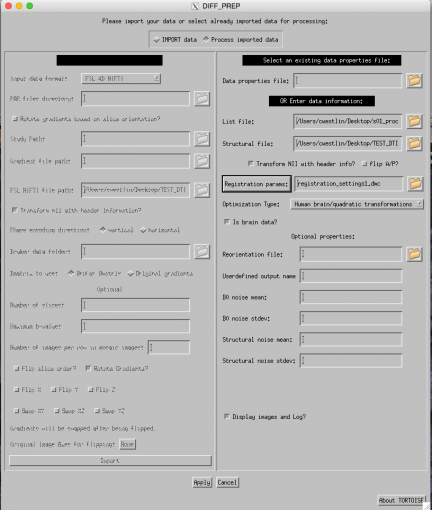
The bulk of our DTI analysis will involve the NIH's open source program TORTOISE.
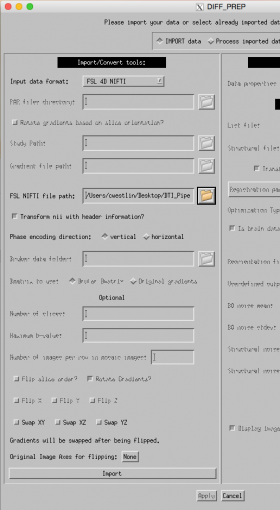
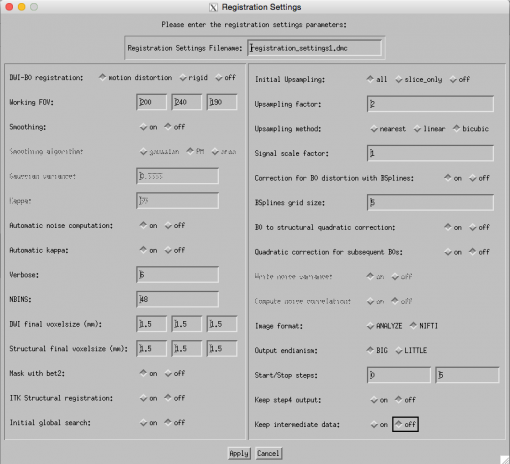
BUT, before we use TORTOISE, we need to prepare our anatomical image for use in this program.
It is recommended that you use a T2-weighted anatomical image in TORTOISE because it has similar characteristics to diffusion data. However, we are using a T1-weighted image for this tutorial because we did not collect a T2 image for this dataset.
Through consulation with a TORTOISE developer, we came up with a way to utilize a T1 image when lacking a T2. This involves separating brain and non-brain regions, attenuating the non-brain regions, and then putting the brain regions back together with the attenuated non-brain regions to produce a final attenuated-T1.
If you are using a T2 image rather than following along with our example dataset, you can skip the following steps below and proceed with Step 2.
Otherwise, perform the following steps to produce an attenuated-T1 image for use with TORTOISE.
First, we will use FSL's fslreorient2std command to re-orient our anatomical image to the orientation of the MNI152 standard template. To do so, run the following command for each participant's T1 image:
fslreorient2std s01_T1.nii s01_T1_std.nii
Next, we will remove non-brain tissue from our T1 image using FSL's BET tool by running the following command for each participant's T1 image:
bet s01_T1_std.nii s01_T1_bet.nii.gz -B -f 0.4 -g 0
NOTE: This step will take ~10 mins-15**
Then, we will attenuate the non-brain regions of the T1 and add the attenuated non-brain regions back to the brain regions to produce a final attenuated-T1. To do so, run the following command:
fslmaths s01_T1_std.nii -sub s01_T1_bet.nii.gz -div 10 -add s01_T1_bet.nii.gz s01_T1_att.nii
NOTE: This will create a zipped file named s01_T1_att.nii.gz. Before continuing on with the next steps, unzip this file by typing this command: gunzip s01_T1_att.nii.gz